In the field of biological sequence modeling, advances in deep learning technology are impressive, but the high computing needs and dependence on large data sets have troubled many researchers. Recently, a research team from MIT, Harvard University and Carnegie Mellon University has launched a novel biological sequence modeling method called Lyra. Not only does this method significantly reduce the parameters to only one-twelve thousand of the traditional model, but it can also be trained using two GPUs in just two hours, greatly improving the efficiency of the model.
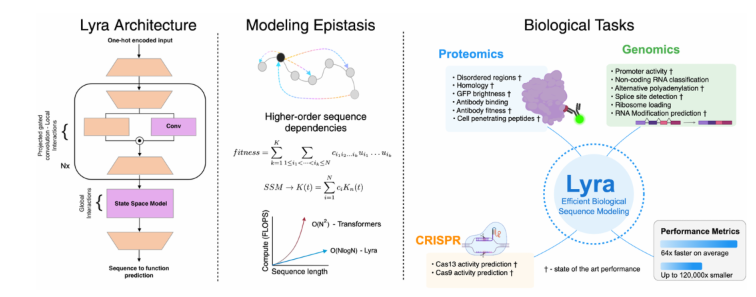
Lyra’s design is inspired by the epistaxis in biology (i.e., the interaction between mutations within sequences), which effectively understands the relationship between biological sequences and their functions through a secondary architecture. This new model has excellent performance in more than 100 biological tasks, including protein fitness prediction, RNA functional analysis, and CRISPR design, and even achieves the best performance of current technology (SOTA) in some key applications.
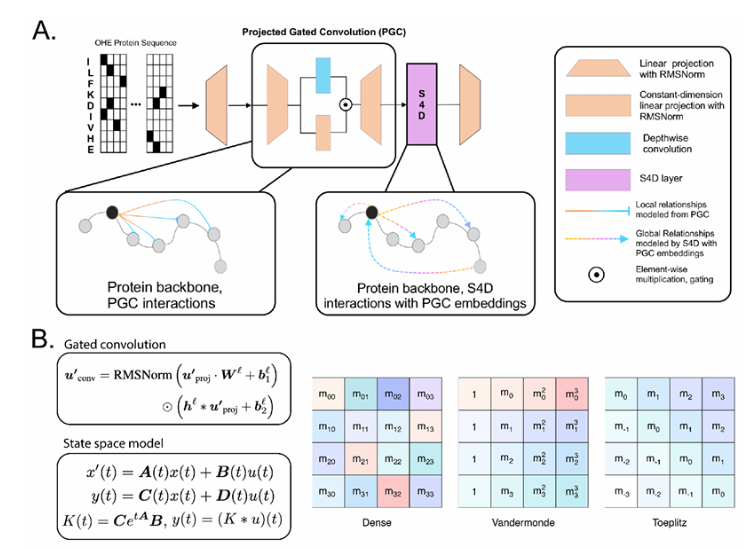
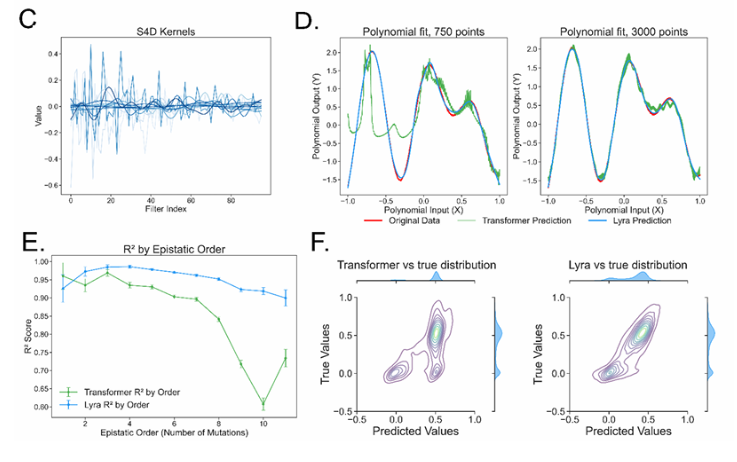
Compared with traditional convolutional neural network (CNN) and Transformer models, Lyra's inference speed is 64.18 times, while significantly reducing parameter requirements. This is thanks to its innovative hybrid model structure, where Lyra combines state space model (SSM) and projection gating convolution (PGC) to capture local and global dependencies in biological sequences. SSM efficiently model global relationships through fast Fourier transform (FFT), while PGC focuses on extracting local features. The combination of the two allows Lyra to achieve a good balance between computational efficiency and interpretability.
Lyra's efficiency can not only promote the progress of basic biological research, but may also play an important role in practical applications such as treatment development, pathogen monitoring, and biomanufacturing. The research team hopes that through Lyra, more researchers can model complex biological sequences with limited resources, thereby accelerating the exploration of biological science.